Characterization Intro
TECHNOLOGY FOCUS![[Dividing Line Image]](/mwf/pictures/div.gif)
INTRODUCTION TO THE CHARACTERIZATION OF METALWORKING FLUIDS
PART I
by Pat Williams
Microbial Insights
2340 Stock Creek Blvd.
Rockford, TN 57853
www.microbe.com
February 2001:
INTRODUCTION
Over the past several years concerns have been expressed over the potential health effects associated with chronic exposure to metal working fluids (MWFs). Scientific evidence indicates that workers exposed to MWF aerosols have an increased risk of respiratory and skin disease. A comprehensive, non-culture based methodology has been developed for the assessment of the microbial community present in metalworking fluids. Characterization of the MWF is based on DNA testing and Phospholipid Fatty Acid Analysis (PLFA).
Culture based approaches are limited to isolating and colonizing target bacteria that are able to grow in specified conditions. Standard plating techniques can be time consuming, selective, and fundamentally change microorganisms from their original status. Isolated cultures provide little information about their original condition or role within the sample. To overcome these limitations, analytical methods have been developed to characterize microorganisms directly from their environment.
Advances in instrumentation and procedures have led to enormous improvements in the analysis of microbial communities directly. These techniques now offer new opportunity in microbial ecology that was not possible using earlier methods such as direct culturing. A variety of matrixes can be analyzed using these methods including metalworking fluids, sump tank biofilms, and aerosols. Lipid profiling methods are quantitative and can address physiological status questions. Nucleic acids, commonly referred to as DNA, encode information about the organisms identity and potential activities within the MWF itself (including antibiotic resistance). Application of these new analytical techniques can provide a more comprehensive analysis of the microbial communities present in MWFs in a shorter period. These new methods present a real opportunity to monitor the biological populations within MWFs and provide data that are more complete in order to protect the health of workers.
DNA ANALYSIS
Within an organism, the genetic material, commonly known as DNA, determines its identity. During the course of evolution, different regions of the DNA have mutated, or changed. The overall result is that some regions of the DNA are virtually the same between all organisms, while other regions differ between even closely related species. Other regions of the DNA have mutated at intermediate rates, such that they differ between kingdoms, subgroups, or sister genera yet maintain a similar sequence from species to species. By selecting DNA targets that carry detailed taxonomic information, the individual microbes of the target community can be identified.
The favored target in DNA identification for microbes is the small ribosomal sub-unit RNA gene, generally referred to as “16S rDNA” in prokaryotes (bacteria), and “18S or 28S rDNA” in eukaryotes (fungi). The sequence of a 16S-rDNA molecule from an “unknown” organism can always place it within the framework provided by 16S rDNA sequences of cultured organisms, providing a match of the unknown. The strength of the placement is generally dependent on its similarity to known sequences, and the length of the sequence recovered.
Microbial DNA is extracted directly from MWF samples, which then undergo polymerase chain reaction (PCR), an enzyme driven reaction that makes thousands of copies of the 16S, 18S, or 28S rDNA genes from the whole community DNA sample. The process of copying the DNA is known as amplification. The PCR product contains amplified DNA, that are roughly the same length, but with slightly different sequences from the various organisms in the sample. It is through a combination of targeting these similar or conserved regions and the regions with the highly varied sequences that we are able to first amplify all the organisms in the targeted group and then genetically separate and identify them.
These amplified strands of DNA (PCR product) are separated based upon their melting behavior in a polyacrylamide gel through a process called Denaturant Gradient Gel Electrophoresis (DGGE). When this double stranded DNA is subjected to the gel gradient, it partially separates, or melts. As the strands of DNA migrate and separate through the gradient gel, different sequences form bands in varying positions. The intensity of each band is related to the original abundance of DNA and the banding patterns show differences in each MWF sample. Each band can be excised and sequenced for direct identification using either the Ribosomal Database Project (RDP) or Gen Bank.
PLFA is a quantitative method for gathering a significant amount of information about the microbial community present in MWFs. The technique is based on the extraction of “signature” lipid biomarkers from cell membranes of microorganisms. Lipids are macromolecules comprised of an alcohol and a fatty acid that are insoluble in water. They function as the principle component of cellular membranes to control the transport of materials in and out of the cell. These lipid membranes differ for groups of organisms and change in response to growth phases or environmental conditions.
The PLFA methods described in this report allows extraction of these lipids from whole microbial communities directly from samples without having first to isolate and culture the organisms.
Once extracted, lipids are concentrated, separated, and then analyzed by gas chromatography/mass spectroscopy (GC/MS) for identification of each individual compound. A profile of the fatty acids and other lipids is then used to determine the characteristics of the microbial community. Specifically, the analysis provides quantitative insight into three important attributes: viable biomass, community structure, and metabolic status.
The total viable microbial biomass can be determined by the amount of Phospholipid ester-linked fatty acids (PLFA) detected in a sample. Viable microbes have an intact membrane that contains phospholipids. Cellular enzymes hydrolyze and release the phosphate group from the fatty acids within minutes to hours of cell death; therefore analysis of PLFA provides a measure of cells with intact membranes.
Specific patterns of PLFA can indicate physiological stress. Exposure to toxic environments can lead to mini-cell formation, a relative increase in specific PLFAs. Calculations of certain PLFA ratios indicate the sate of growth and the exposure of the microbes to environmental stress, such as toxicity or starvation. In Gram Negative bacteria, monenoic fatty acids are converted to cyclopropyl fatty acids as microbes move from a long to a stationary phase of growth. Gram Negative bacteria also generate trans fatty acids to minimize the permeability of their cell membranes as protection against changes in the environment such as toxicity or starvation. The relative increase in trans fatty acids shows the gram-negative bacteria reacting to their environment.
PROJECT SUMMARY
Three air samples and two bulk samples were characterized by PLFA analysis. Results from the PLFA analysis, showed that biomass content increased in both the air and bulk samples. The indoor air samples contained moderately diverse bacterial populations primarily composed of Gram-negative bacteria. The bulk samples contained relatively simple microbial populations with the dirty sample primarily composed of Gram-negative bacteria and the clean sample primarily composed of eukaryotes.
The DGGE results showed that species composition was different between the dirty and clean metal working fluids. Additional air samples collected for DNA showed that several organisms within the clean and dirty fluids were present within the air.
R
Phospholipid fatty acids (PLFA) are found within the membranes of all living cells but decompose quickly upon cell death. Thus, measuring PLFA content provides a quantitative measure of the viable microbial biomass present (1-2).
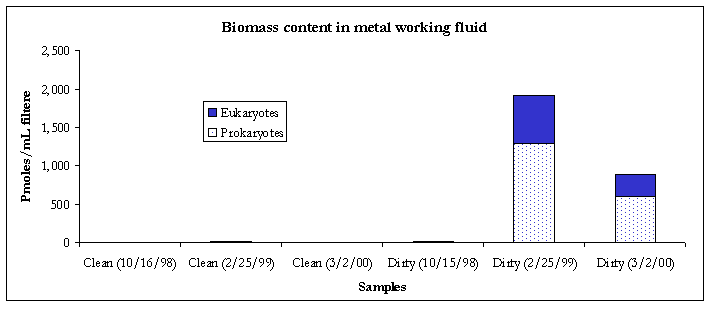
Figure 1. Biomass content.
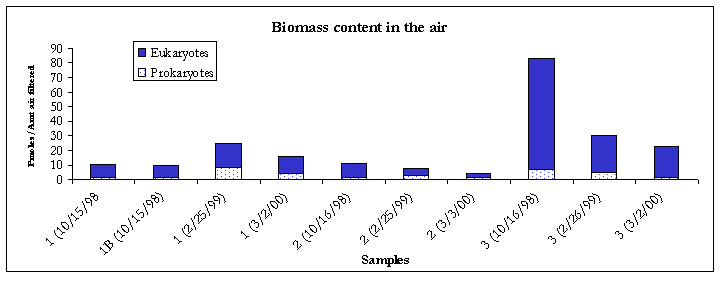
Figure 2. Biomass content.
Compared to the 2/26/99 event, results from the 3/2/00 sampling event showed that biomass content had decreased in the dirty metal working fluid. Within the air samples, biomass levels also showed a decrease at all three locations (Figure 1).
Community Composition
To relate the complex mixture of PLFA to the organisms present, a structural group interpretation is employed (3-5). In some cases these associations are so strong that fatty acid biomarkers have been identified for particular organisms.
Table 1. PLFA Structure Groups.
|
PLFA Structural Group |
General classification |
|
Monoenoics (Monos) |
Found in Gram-negative bacteria, which are fast growing, utilize many carbon sources, and adapt quickly to a variety of environments. |
|
Terminally Branched Saturated (TerBrSats) |
Representative of Gram-positive bacteria, but may also be found in the cell membranes of some Gram-negative bacteria. |
|
Branched Monoenoic (BrMonos) |
Commonly found in the cell membranes of obligate anaerobes such as sulfate or iron reducing bacteria |
|
Mid-Chain Branched Saturated (MidBrSats) |
Common in Actinomycete, sulfate reducing bacteria and certain Gram-positive bacteria. |
|
Normal Saturated |
Found in both the prokaryotic and eukaryotic kingdoms. |
|
Eukaryotes |
Found in organisms such as fungi, protozoa, algae, higher plants and animals. |
Indoor air samples
The indoor air samples contained moderately diverse bacterial populations primarily composed of monoenoic PLFA. Generally monoenoic PLFA are indicative of the presence of Gram-negative bacteria, which are fast growing, utilize many carbon sources and adapt quickly to variety of habitats. Where indoor air contamination is present, monoenoic PLFA will generally comprise at least 8% of the total fatty acids detected. Terminally branched saturate PLFA were detected in the #2 (2.2%) and #3 (0.6%) air samples. Terminally branched saturate PLFA are representative of Gram-positive bacteria but may also be found in the cell membranes of many sulfate-reducing bacteria. However, no branched monoenoic PLFA, which are usually found in the cell membranes of obligate anaerobes such as sulfate reducing bacteria, were detected. Therefore, the terminally branched saturate PLFA were deemed to be mainly from Gram positive in bacteria. Normal saturate PLFA were detected in all the indoor air samples. Normal saturate PLFA are found throughout the prokaryote and eukaryote kingdoms so little conclusion could be drawn as to their exact origin.
Bulk Samples
The 3/2/00 bulk samples contained relatively simple microbial communities with clean sample primarily composed of eukaryotes and dirty sample primarily composed of monoenoic PLFA. There are several biomarkers for different microeukaryotic organisms. The biomarker 18:2w6 which is prominent in fungi, but is also found in algae, protozoa, higher plants and animals was detected in both bulk samples (clean; 11.7% and dirty; 0.7%). Monoenoic PLFA are typically found in Gram negative bacteria, which are fast growing, utilized many carbon sources and adapt quickly to a variety of environments.
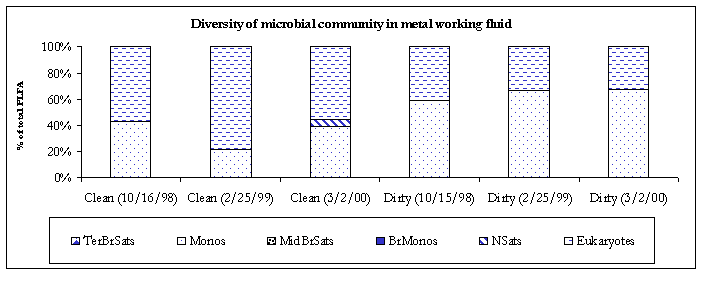
Figure 3. Diversity of the microbial communities.
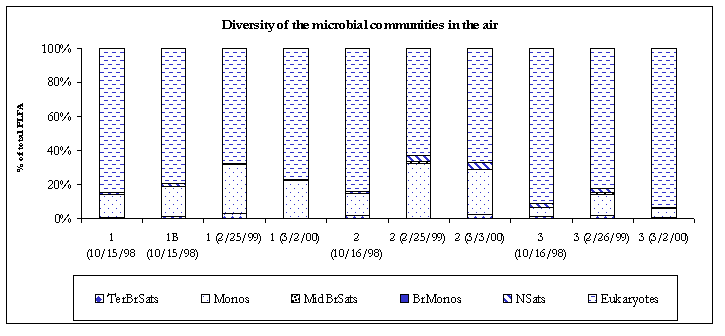
Figure 4. Diversity of the microbial communities.
Metabolic status
The lipid composition of microorganisms is a product of metabolic activity and thus reflects the phenotypic response of the organisms to their environment. The physiological status of Gram-negative communities can be assessed from ratios of different monoenoic biomarkers. Specifically, 16:1w7c and 18:1w7c are converted to cyclopropyl fatty acids (cy17:0 & cy19:0) as microbes move from a log to a stationary phase of growth (i.e. slowing of growth). This change is expressed in the two ratios cy17:0/16:1w7c and cy19:0/18:1w7c, which may vary from organism to organism or environment to environment but usually will fall within the range of 0.1 (log phase) to 5.0 (stationary phase) when summed. This ratio is inversely proportional to the turnover rate, i.e. a lower ratio infers a higher turnover rate. An increase in cyclopropyl formation also has been associated with anaerobic metabolism. Gram-negative bacteria also generate trans fatty acids to minimize the permeability of their cellular membranes as adaptation to a more hostile environment. Ratios (16:1w7t/16:1w7c and 18:1w7t/18:1w7c) greater than 0.1 (when summed) have been shown to indicate an adaptation to a toxic or stressful environment resulting in decreased cellular membrane fluidity, which decreases membrane permeability (6).
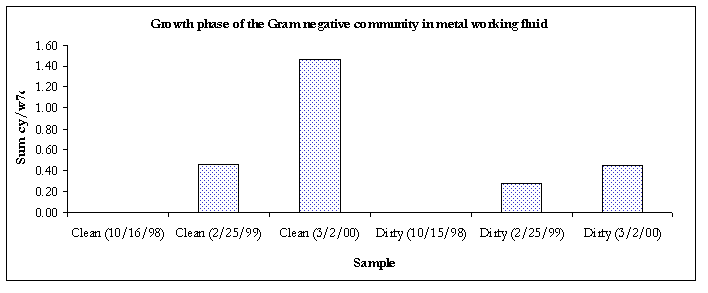
Figure 5. Growth phase of the Gram-negative communities.
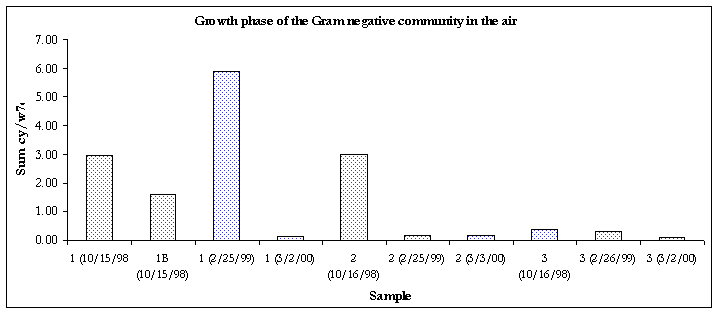
Figure 6. Growth phase of the Gram negative communities
The Gram-negative communities in samples taken from 3/2/00 were in the stationary phase of growth.
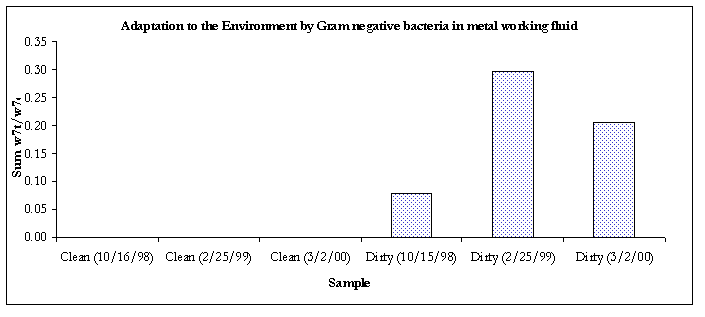
Figure 7. Evidence of adaptation by decreased membrane permeability.
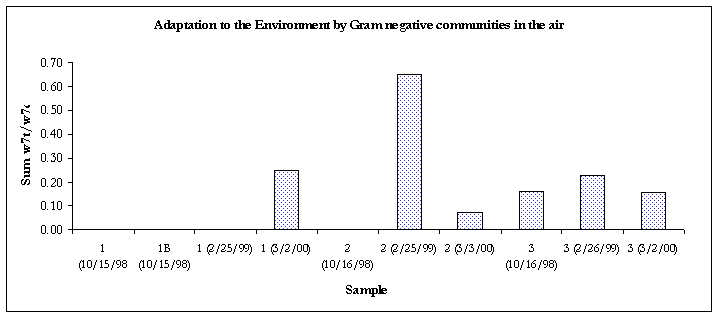
Figure 8. Evidence of adaptation by decreased membrane permeability.
Except for the air sample #2 and the clean bulk sample, all of the Gram-negative communities from samples taken from 3/2/00 were showing signs of decreased membrane permeability, a bacterial response to environmental stress.
Bacterial Composition
The DGGE approach directly determines the species composition of complex microbial populations based on the amplification of 16S rDNA fragments. DNA fragments of the same length but with different base pair sequences are separated based on their melting behavior in a polyacrylamide gel containing a linearly increasing gradient of denaturant. The banding patterns and relative intensities of the recovered bands provide a measure of change in the community. Dominant species, which constitute at least 1% of the total bacterial community, can be excised and sequenced. Sequence analysis of individual bands is used to infer the identity of the source organism based upon database searches and phylogenetic methods (7).
CONCLUSION
The bacterial communities within two bulk and five air samples were characterized by DGGE analysis. Results from this analysis showed distinct differences between the clean (bulk 2) and dirty (bulk 1) samples. Several of the more prominent bands were present within the air samples. Sequence results are found in table 2. These new analytical methods provide quantitative data on the microbial populations present within MWFs without the time and labor constraints typically associated with culturing. In addition, the data generated by these methods is more useful for the protection of workers and determining the lifetime of the MWF.
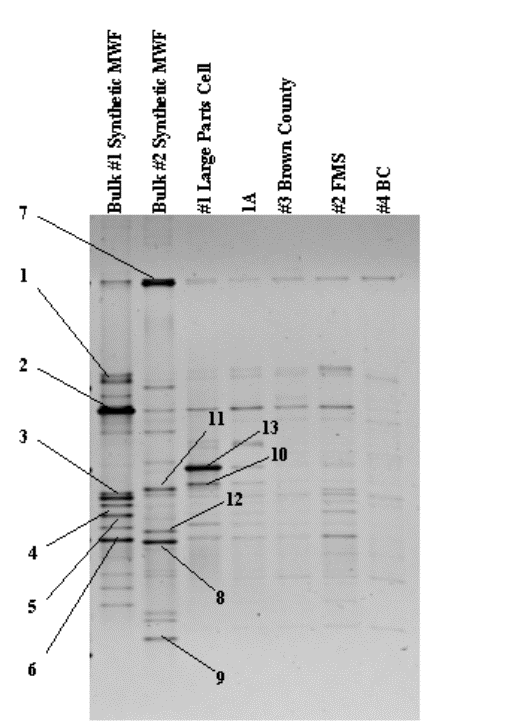
Figure 9. DNA profile of dominant bacterial species.
Table 2. Sequence Results.
|
Band # |
Best Match |
% Match
|
Phylogenetic affiliation |
|
1 |
Pseudomonas sp. |
95% |
Gamma Proteobacteria |
|
2 |
Pseudomonas balearica |
96% |
Gamma Proteobacteria |
|
3 |
Acidovorax avenae |
95% |
Beta Proteobacteria |
|
4 |
Lysobacter antibioticus |
95% |
Gamma Proteobacteria |
|
5 |
Alcaligenes sp. |
97% |
Beta Proteobacteria |
|
6 |
Burkholderia pseudmallei |
97% |
Beta Proteobacteria |
|
7 |
Herbaspirillum seropedicae |
96% |
Beta Proteobacteria |
|
8 |
Bradyrhizobium elkanii |
99% |
Alpha Proteobacteria |
|
9 |
Pedomicrobium americanum |
95% |
Alpha Proteobacteria |
|
10 |
Sphingomonas adhaesiva |
97% |
Alpha Proteobacteria |
|
11 |
Ralstonia picketti |
93% |
Beta Proteobacteria |
|
12 |
Mesorhizobium sp. |
98% |
Alpha Proteobacteria |
|
13 |
Multiple sequences |
- |
- |
References
1. White, D.C., et al. (1998) In situ microbial ecology for quantitative appraisal, monitoring, and risk assessment of pollution remediation in soils, the subsurface, the rhizosphere and in biofilms. J. Microbiol. Methods
2. Balkwill, D.L., et al. (1988) Equivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate and direct counts in subsurface sediments. Microbial Ecol. 16, 73-84.
3. Tunlid, A., White, D.C. (1991) Biochemical analysis of biomass, community structure, nutritional status and metabolic activity of the microbial communities in soil, in Soil Biochemistry, Vol. 7, (Bollag, J.M., Stotzky, G. eds.), pp. 229-262.
4. Dowling, N.J.E., et al. (1986) Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulfate reducers and other sulfide-forming bacteria. J. Gen. Microbiol. 132, 1815-1825.
5. White, D.C., et al. (1980) Nonselective biochemical methods for the determination of fungal mass and community structure in estuarine detrital microflora. Botanica Marina 23, 239-250.
6. Guckert, J.B., et al. (1986) Phospholipid, ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans / cis ratio and proportions of cyclopropyl fatty acids. Appl. Environ. Microbiol. 52, 794-801.
7. Muyzer, G., et al. (1993) Profiling of Complex Microbial Populations by Denaturing Gradient Gel Electrophoresis Analysis of Polymerase Chain Reaction-Amplified Genes Coding for 16S rRNA. Appl. and Env. Microbiol. 59, 695-700.


